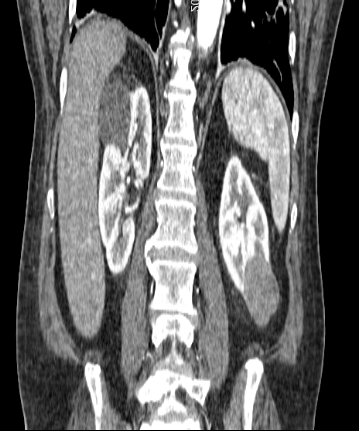
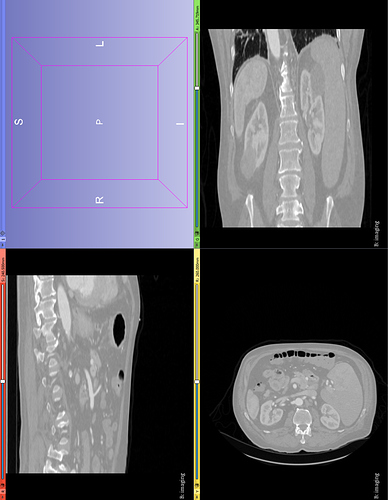
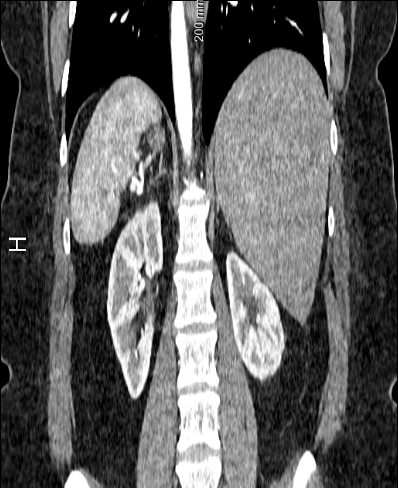
The default spacing values are 1, 0.92, 0.92 for z, x, y directions but the 3D volume looks stretched in the z direction (along Sagittal plane). Setting the z spacing (Sagittal plane) to be 0.5 makes 3D volumes look more real but what is the actual z spacing value?
Thanks,
Yanling