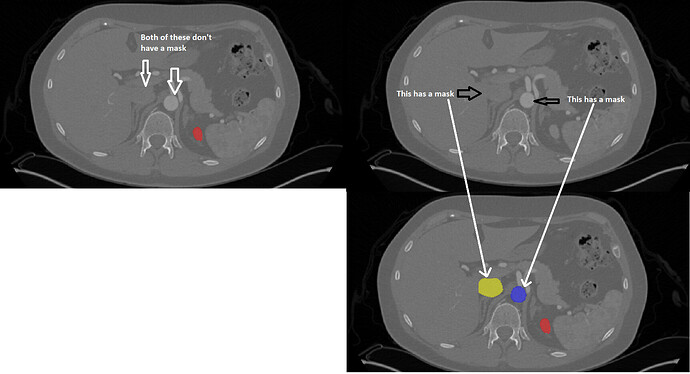
Thanks for bringing this up. In order to focus our annotation efforts near to the kidney, we bounded our artery and vein annotations above and below at consistent anatomical landmarks:
- On the superior side: the IVC and aorta are bounded above by the axial slice at which the celiac artery bifurcates from the aorta – shown above the aorta in your top right image
- On the inferior side: the IVC, aorta, and ureters are bounded below by the axial slice at which the aorta bifurcates into the common iliac arteries.
The kidney, tumor, and cyst regions have no such boundaries, which is why the kidney mask is still present in your top left slice.
Since this intent is consistent in our training data, we expect the nets to be able to identify these landmarks fairly easily and make predictions accordingly. That said, if this issue causes trouble for our internal baselines, we might revisit this point with either a modified metric or by cropping the imaging. We’re also open to suggestions